Elisabetta Mereu
Group Leader of the Cellular Systems Genomics Group
Josep Carreras Research Institute (Barcelona, Spain)
Biography
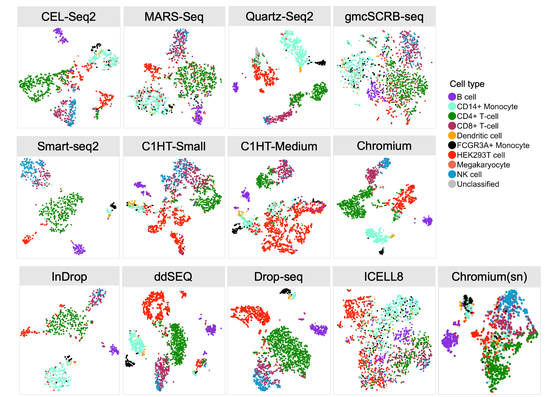
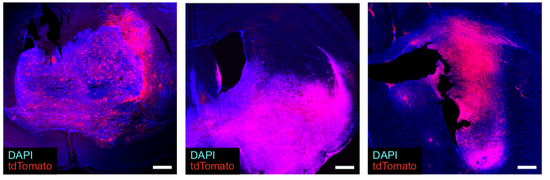
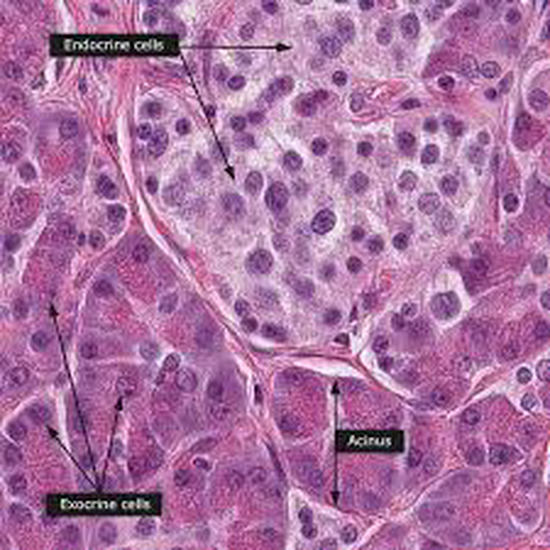
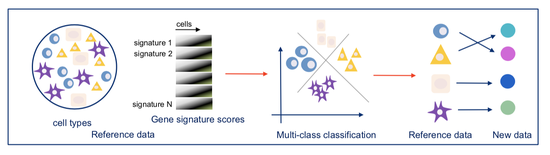
From 2021 I’m Group Leader of the Cellular Systems Genomics Group at the Josep Carreras Institute, in Barcelona. The group focus is to define the spatial-temporal organization of complex tissues in health and disease. We are particularly interested in inflammatory disorders and autoimmune diseases. By adopting a single-cell perspective, we will provide new machine learning tools and computational methods in order to unlock key regulatory mechanisms hidden in large-scale multi-omics datasets. During my postdoc at the National Centre of Genomic Analysis of Spain, I worked as computational scientist at the Single Cell Genomics Team, in which I gained experience on the analysis of multi-modal data that are from individual cells. Being in a high throughput sequencing center allowed me to be involved in a wide range of projects in collaboration with local and international research groups. As part of the Human Cell Atlas consortium, I work on the benchmarking of scRNA-seq protocols and the integration of multi-modality sequencing data (e.g. sc/scRNA-seq, bulk-RNA-seq, spatial transcriptomics and scMNT-seq) in order to quantify reproducibility and complementarity across these methods. I’m also part of the Expression and Spatial analysis Pancreas Atlas Consortium Europe (ESPACE) for the generation of the first version of the Human Cell Atlas of the Pancreas.
Interests
- Single-cell and Spatial Genomics
- Computational biology
- Inflammation and Autoimmunity
- Cancer Genomics
Education
PhD in Statistical Genetics, 2015
University of Cagliari
MSc in Mathematics, 2009
University of Cagliari
BSc in Mathematics, 2007
University of Cagliari